
Imprinted genes, a fundamentally epigenetic phenomena, are genes which show genotype-independent parent-of-origin effects. Genomic imprinting is a good example of an epigenetic effect because a key component of the environment (i.e., whether the gene was transmitted from sperm or egg) leaves a functional but reversible mark on DNA. Once the offspring produces its own gametes (i.e., sperm or egg), parent-of-origin epigenetic marks are normally reset. In the case of imprinted genes, the previous parental environment is trans-generationally affecting the next generation without altering the underlying nucleotide sequence, see Figure 1. It is well-known that these rare genes (~1-2 of human genome) have large effects on growth, brain and behaviour in humans and other animals.
Figure 1. (A) Illustration of genomic imprinting compared to (B) X-inactivation. Both are epigenetic phenomenon.
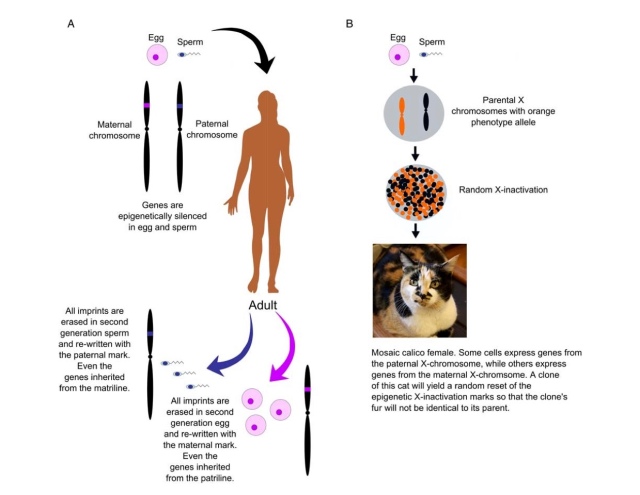
The best evolutionary explanation for the existence of imprinted genes in mammals is kinship theory. Kinship theory, proposed by Professor David Haig, hypothesises that in species with multiple paternity (or relatedness asymmetries due to other factors), genes of matrilineal and patrilineal descent have differing inclusive fitness interests in terms of offspring growth and behaviour. Specifically, paternal genes within offspring (i.e., patrigenes to distinguish between genes within the father) are expected to increase costs on matrilineal inclusive fitness where it benefits themselves, whilst matrigenes are expected to minimise these costs. Perhaps the most dramatic developmental example of differing interests between parental genomes can be seen in Figure 2 below.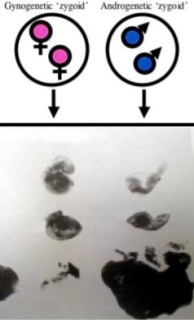
Figure 2. Germ cell pronuclear transplant experiments in mice (e.g., performed by Surani, McGarth and Solter in the 1980’s) demonstrated a possible ‘tug of war’ over growth between sperm- and egg-derived nuclear genomes during prenatal development. Development in the absence of a sperm-derived genome (left column) shows development of the embryo proper but failed development of the trophoblast. Development in the absence of an egg-derived genome (right column) shows failed development of the embryo proper but trophoblast growth. Figure modified from Nature.
In my 2015 paper published online first for the British Journal of Sports Medicine, I systematically investigated whether or not imprinted genes, despite their rarity in the human genome, are implicated in human skeletal muscle gene networks and secondly in exercise-associated DNA methylation change.
Link to the paper for free download.
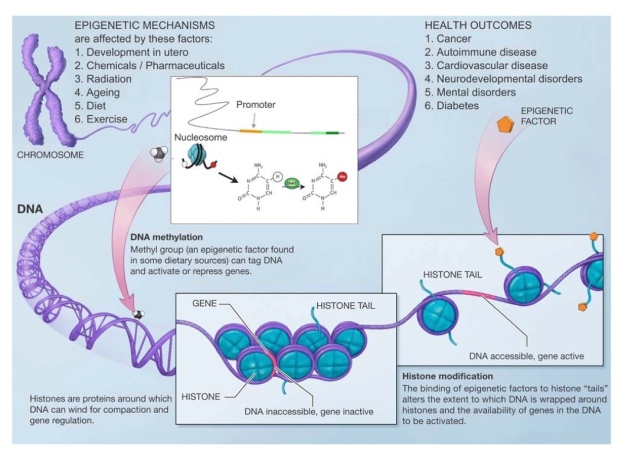
To better understand the paper some background information is needed. DNA methylation is the addition of methyl groups to a region of DNA which can affect gene expression. For example, the addition of methyl groups at the promoter region of DNA can normally cause reduced gene expression. See Figure 3 for an illustration of DNA methylation, its mechanisms, causes and consequences.
Figure 3. Factors affecting epigenetic mechanisms and health outcomes. Image modified from the National Institutes of Health, Benjamin I. Laufer and Forluvoft.
What is known about exercise epigenetics and genomic imprinting
Recent evidence suggests that adult physical activity acutely (i.e., seemingly temporarily) modifies regions of the human epigenome. Specifically, DNA methylation, an important gene expression regulator and correlate of diverse disease states, is temporarily altered by physical activity. No systematic review has been conducted to elucidate these associations and whether or not imprinted genes are implicated.
What Brown (2015) adds to the field of exercise epigenetics
Brown (2015) isolates imprinted genes, known to be important for health and disease, as correlates of skeletal muscle growth and targets of exercise. Amongst older people, there were significant benefits from exercise in terms of the possible adaptive epigenetic regulation of tumour suppressor genes. These findings may help explain why exercise appears to have beneficial effects on cancer and provides a list of epigenetic candidate loci worthy of further empirical scrutiny.
Brown’s (2015) paper has implications for multiple fields. Below is a brief outline of the importance of this paper for different fields and an outline of where caution is required regarding methodology and interpretation.
Importance for medicine
I provide novel analyses and explanations for the inverse relationship between exercise and cancer in humans. Specifically, epigenetic mechanisms (e.g., DNA methylation of gene networks some of which are regulated by microRNAs) appear sensitive to physical exercise. Exercise-associated DNA methylation change amongst older people were particularly robust, especially for gene networks important for tumour suppression. Given that simple inexpensive interventions (e.g., walking) appear to have robust associations with short-term DNA-methylation change amongst tumour suppressors, these interventions could become important for medical practice. Two recent studies are of particular relevance here. For example, Zeng et al (2012) showed that among breast cancer sufferers, DNA methylation of L3MBTL1 (an imprinted and possible tumour sup- pressor gene) was altered due to exercise (e.g., brisk walking on a treadmill). Importantly, probability of survival was increased after the intervention. A second study by Maeda et al (2011) amongst older people with cerebrovascular disease suggested that physical function improvements during hospitalisation covary with sub-telomeric methylation of long telomeres. Studies such as these are just the beginning and much more mechanistic research is required to fully understand these possible epigenetic mechanisms mediating the association between exercise and positive health outcomes amongst older people. An applied goal would be to adaptively decouple chronological and epigenetic age by using exercise interventions. The analyses presented in Brown (2015) suggest that exercise-associated DNA methylation change could reduce epigenetic age. Given Professor Steve Horvath’s ground-breaking research on epigenetic age, the benefits of exercise on the epigenome in multiple tissues may be readily quantifiable using a quasi-experimental approach.
Importance for evolutionary biology
The fact that exercise-associated DNA methylation change was stronger amongst older compared with younger people indicates that exercise could alter an organism’s ‘epigenetic age’ by warding off senescence. Why would exercise have more profound effects on older compared with younger epigenomes? One possibility is that as organisms age, epigenetic errors accumulate, and because there are more to correct or reset, older people experience greater (and more positive) DNA methylation change upon exercise intervention compared with younger people (who have experienced fewer epigenetic errors). This parsimonious explanation is similar to regression to the mean due to a random rebound from a ceiling effect amongst the aged. However, since physically active grandparents were probably a characteristic of the majority of human evolution, the pronounced age effect could also be an example of an age-dependent adaptive epigenetic response to antagonistic pleiotropy. Antagonistic pleiotropy is the hypothesis proposed by the late George C. Williams that genes with multiple effects can be beneficial at younger ages and costly later in life. In the context of growth effects, older individuals’ tumour suppressor genes may become demethylated (possibly more expressed), but growth-related loci become methylated (presumably less expressed). Thus, rebuilding of new tissue would be costly for older relative to younger people. To reiterate, stem cells and tissue regeneration adaptive gene complexes in younger people may well be particularly costly amongst older organisms due to cancer proliferation as we age. The inclusive fitness cost of cancer among older people would be lack of ability to cross-generationally invest in younger relatives. It is important to note that a reverse antagonistic pleiotropic effect may be operating. Specifically, growth suppression among younger individuals may be particularly costly (e.g., inability to regenerate or develop secondary sexual characteristics) relative to older post-reproductive individuals. These ideas are speculative, but from an evolutionary perspective it is tantalising that there appear to be seemingly adaptive epigenetic responses to antagonistic pleiotropy as we age.
In utero transgenerational epigenetics?
Beyond repeated exposure of physical exercise across the lifespan being important for a healthy epigenome, in utero maternal epigenetic effects are likely to be important. There is recent work in mice showing that maternal exercise during pregnancy can reverse the deleterious epigenetic effects of poor maternal diet on newborn pups’ Pgc-1a (Laker et al 2012) . The beneficial effects of maternal exercise during pregnancy on Pgc-1a DNA methylation levels in the next generation suggest that a transgenerational mechanism exists for long-lasting epigenetic changes and is consistent with a foetal origins of disease approach (Chalk & Brown, 2014). There is no evidence of transgenerational DNA methylation effects of exercise in humans (Chalk & Brown, 2014). However, it is note- worthy from the meta-analysis that GRB10 (γ isoform)—maternally expressed in human foetal skeletal muscle (Blagitko et al, 2000)—showed greater exercise-associated DNA methylation change among those without a type 2 diabetes family history. Relaxation of the growth-inhibiting effects of maternal genes could be dependent on genetic, ecological conditions or foetal exposure to maternal exercise. For example, individuals from families exhibiting more sedentary behaviour (i.e., characterised by a history of type 2 diabetes) have less silencing of maternally expressed GRB10 in skeletal muscle. Such differentially epigenetic responses of GRB10’s γ isoform depending on a family history of type 2 diabetes would be extremely interesting if reliable. A powerful interface between family history and offspring epigenetics could be signalled during gestation. Specifically, maternal physical activity during gestation could produce dose-dependent epigenetic responses in offspring (Chalk & Brown, 2014). For a brief review of phenotypic changes in human offspring from maternal exercise during pregnancy see the following blog post. It remains to be seen whether or not level of physical activity among pregnant human mothers leaves epigenetic traces on the DNA of their children.
Importance for psychology
Psychology is the study of brain and behaviour in humans and other animals. Brown (2015) suggests that exercise behaviour may have beneficial epigenetic effects on cancer-related gene activity. The idea that behaviour can be a positive intervention strategy has a long history in the social and behavioural sciences. Human studies in exercise epigenetics are required not only because of the impact on health of ageing populations, but also because key epigenetic elements (i.e., imprinted genes) responsible for regulating adiposity, energy expenditure, glucose homoeostasis and hunger are differentially imprinted (or read differently) across species.
Cautions
It is worth noting that exercise-associated DNA methylation changes for imprinted genes occurred only in studies where participants were exposed to longer term exercise (i.e., 6 months) as opposed to short bouts of exercise. This interpretation should be taken with caution as it is biased by the fact that fewer genes were studied in the acute study by Barrès et al (2012). Specifically, Barrès et al (2012) selected genes from a previous study of DNA methylation in patients with type 2 diabetes, while the long-term exercise studies revealing imprinted genes (Brown, 2015) screened many more genes using Illumina’s Infinium HumanMethylation450 BeadChip (San Diego, California, USA). This could lead to false positives despite the fact that statistical corrections were employed in the analyses.
Future work needs to be conducted to test whether or not imprinted Differentially Methylated Regions (DMRs) are modified by exercise. Once again, given the relevance of imprinted genes for human cancers, one long-standing conundrum in medicine could be resolved. Specifically, why does exercise treatment appear to reduce the incidence of cancers? One answer is that tumour suppressor genes are ‘reactivated’ at promoters after long-term exercise treatments and there is a corresponding reduction in DNA methylation. Given these possible medical benefits, future research should look at the relationship between exercise stress and regulation of imprinted genes in order to understand more fully the underlying mechanisms. However, despite using two different methods (ie, g-Profiler gene ontology network analysis vs meta-analysis), multiple imprinted loci appear to be missing. This raises the distinct possibility that additional imprinted genes will be found to be associated with skeletal muscle and exercise adaptation in humans. For example, in newborns with transient neonatal diabetes, the loss of an epigenetic mark at the TNDM locus on chromosome 6q24 in the mesodermal lineage causes abdominal muscle hypoplasia, the so-called prune belly sequence.When Laborie et al (2010) investigated a family with prune belly syndrome that included one discordant set of MZ twins, the twin with prune belly (relative to the normal co-twin) had extensive loss of methylation at the TNDM locus, as well as at the following imprinted loci: IGF2R, DIRAS3 and PEG1. Future work in humans and other animals should be able to develop a more comprehensive list of imprinted genes regulating skeletal muscle and associated with exercise. One reason that some imprinted genes may be missed from these analyses could be due to the fact that imprinted genes are often involved in neural systems,which, unlike skeletal muscle, cannot be extracted from healthy human participants.
Notes: Your feedback regarding this paper and ideas are greatly appreciated. Please do get in touch with any questions, suggestions or criticisms so that I can improve these ideas and research. Link to the paper for free download. Below is the abstract for a brief summary of the approach and findings.
Exercise-associated DNA methylation change in skeletal muscle and the importance of imprinted genes: a bioinformatics meta-analysis
ABSTRACT
Background Epigenetics is the study of processes— beyond DNA sequence alteration—producing heritable characteristics. For example, DNA methylation modifies gene expression without altering the nucleotide sequence. A well-studied DNA methylation-based phenomenon is genomic imprinting (ie, genotype- independent parent-of-origin effects).
Objective We aimed to elucidate: (1) the effect of exercise on DNA methylation and (2) the role of imprinted genes in skeletal muscle gene networks
(ie, gene group functional profiling analyses).
Design Gene ontology (ie, gene product elucidation)/ meta-analysis.
Data sources 26 skeletal muscle and 86 imprinted genes were subjected to g:Profiler ontology analysis. Meta-analysis assessed exercise-associated DNA methylation change.
Data extraction g:Profiler found four muscle gene networks with imprinted loci. Meta-analysis identified 16 articles (387 genes/1580 individuals) associated with exercise. Age, method, sample size, sex and tissue variation could elevate effect size bias.
Data synthesis Only skeletal muscle gene networks including imprinted genes were reported. Exercise- associated effect sizes were calculated by gene. Age, method, sample size, sex and tissue variation were moderators.
Results Six imprinted loci (RB1, MEG3, UBE3A, PLAGL1, SGCE, INS) were important for muscle gene networks, while meta-analysis uncovered five exercise- associated imprinted loci (KCNQ1, MEG3, GRB10, L3MBTL1, PLAGL1). DNA methylation decreased with exercise (60% of loci). Exercise-associated DNA methylation change was stronger among older people (ie, age accounted for 30% of the variation). Among older people, genes exhibiting DNA methylation decreases were part of a microRNA-regulated gene network functioning to suppress cancer.
Conclusions Imprinted genes were identified in skeletal muscle gene networks and exercise-associated DNA methylation change. Exercise-associated DNA methylation modification could rewind the ‘epigenetic clock’ as we age.
Trial registration number CRD42014009800.

 This E.T. the Extra-Terrestrial looking 3D scanner (pictured above) was a breakthrough. However, it led to a series of scientific problems or challenges. The first was finding landmarks that were repeatable across different people doing the multiple measurements of the same face. That took several years and multiple research assistants. Along the way it became apparent that some people were more capable at 3D measurement than others. The second issue was finding software and learning a workflow that could yield useful 3D measurements of fluctuating asymmetry and demarcating it from directional asymmetry. I found two pieces of free and wonderful software which allowed me to: (a) place landmarks of 3D specimens (Landmark from Evolutionary Morphing folks at the Institute of Data Analysis and Visualization at The University of California, Davis); and (b) import and measure asymmetries and shape variation (MorphoJ from the Klingenberg Lab at the University of Manchester). It did take time to learn the software and be able to teach undergraduate students how to use it effectively. But we are now there and our first paper is ready for public consumption.
This E.T. the Extra-Terrestrial looking 3D scanner (pictured above) was a breakthrough. However, it led to a series of scientific problems or challenges. The first was finding landmarks that were repeatable across different people doing the multiple measurements of the same face. That took several years and multiple research assistants. Along the way it became apparent that some people were more capable at 3D measurement than others. The second issue was finding software and learning a workflow that could yield useful 3D measurements of fluctuating asymmetry and demarcating it from directional asymmetry. I found two pieces of free and wonderful software which allowed me to: (a) place landmarks of 3D specimens (Landmark from Evolutionary Morphing folks at the Institute of Data Analysis and Visualization at The University of California, Davis); and (b) import and measure asymmetries and shape variation (MorphoJ from the Klingenberg Lab at the University of Manchester). It did take time to learn the software and be able to teach undergraduate students how to use it effectively. But we are now there and our first paper is ready for public consumption.







 e IBANGS 17th Annual Genes, Brain and Behavior (GBB) Meeting provides an excellent opportunity for those involved in research in the fields of behavioural genetics and neurogenetics to present their latest findings, to learn about new research developments, and to interact with others with similar interests. The 2015 GBB meeting will be held on the campus of Uppsala University, the oldest university in Scandinavia (est. 1477), at the Evolutionary Biology Centre.
e IBANGS 17th Annual Genes, Brain and Behavior (GBB) Meeting provides an excellent opportunity for those involved in research in the fields of behavioural genetics and neurogenetics to present their latest findings, to learn about new research developments, and to interact with others with similar interests. The 2015 GBB meeting will be held on the campus of Uppsala University, the oldest university in Scandinavia (est. 1477), at the Evolutionary Biology Centre. Overview
Overview


