Synthesis, in-Vitro Cytotoxicity, Molecular Docking of Few Quinazolinone Incorporated Naphthyl Chalcones: As Potential Dual Targeting Anticancer Agents to Treat Lung Cancer and Colorectal Cancer
Praveen Kumar Arora1* , Sushil Kumar2, Sandeep Kumar Bansal3
, Sushil Kumar2, Sandeep Kumar Bansal3 , Prabodh Chander Sharma4
, Prabodh Chander Sharma4
1Faculty of Pharmacy, IFTM University, Moradabad - 244001, Uttar Pradesh, India.
2School of Pharmaceutical Sciences, Faculty of Pharmacy, IFTM University, Moradabad - 244001, Uttar Pradesh, India.
3Ram-Eesh Institute of Vocational and Technical Education, Greater Noida - 201310, Uttar Pradesh, India.
4School of Pharmaceutical Sciences, Delhi Pharmaceutical Sciences and Research University, New Delhi 110017, India.
Corresponding Author E-mail: procarora@gmail.com
DOI : http://dx.doi.org/10.13005/ojc/390202
Article Received on : 28 Dec 2022
Article Accepted on : 13 Apr 2023
Article Published : 21 Apr 2023
Reviewed by: Dr. Shaheen Begum
Second Review by: Dr. Parvati Sinha
Final Approval by: Dr. S. A. Iqbal
The present study is an effort to explore some low molecular weight chemical entities quinazolinone incorporated naphthyl chalcones for their cytotoxic potential and, that can act smartly by inhibiting the mutated molecular targets EGFR (T790M mutation; PDB Id: 5Y9T), and mutated K-RAS(G12D mutation; PDB Id: 4EPT). The in-vitro cytotoxic studies were done by the MTT assay method. For the lung cancer cell lines (A549), N1-N4 were found as more potent than the reference erlotinib (IC50:44.4μg/ml), and among them, the most potent compound is N3 (IC50:11.29 μg/ml). Against the colorectal cancer cell lines (Caco2), the same compound N3 was found as most potent (IC50:10.79 μg/ml). Molecular docking by autodock-4 revealed that all the title compounds have high affinity for both the molecular targets, as they have high negative binding energies. The inhibition constants obtained in docking studies are in nanomoles, The title compounds can be used as a template for developing more potent, selective and dual targeted drugs to treat lung cancer and colorectal cancer.
KEYWORDS:Anticancer agents; Cytotoxicity; Chalcones; Docking; EGFR inhibitors; K-RAS inhibitors; Quinazolinone
Download this article as:| Copy the following to cite this article: Arora P. K, Kumar S, Bansal S. K, Sharma P. C. Synthesis, in-Vitro Cytotoxicity, Molecular Docking of Few Quinazolinone Incorporated Naphthyl Chalcones: As Potential Dual Targeting Anticancer Agents to Treat Lung Cancer and Colorectal Cancer. Orient J Chem 2023;39(2). |
| Copy the following to cite this URL: Arora P. K, Kumar S, Bansal S. K, Sharma P. C. Synthesis, in-Vitro Cytotoxicity, Molecular Docking of Few Quinazolinone Incorporated Naphthyl Chalcones: As Potential Dual Targeting Anticancer Agents to Treat Lung Cancer and Colorectal Cancer. Orient J Chem 2023;39(2). Available from: https://bit.ly/3N1Op2x |
Introduction
Cancer is one of the deadliest diseases worldwide, one of the most typical malignancies detected in both men and women is lung cancer, followed by colorectal cancer1,2. The analysis of the literature revealed the quinazolinone/quinazoline scaffold’s significant therapeutic potential. Drugs like Raltitrexed, which is used to treat cancer, methaqualone, which has sedative properties, proquazone, which is an NSAID, and albaconazole, which has antifungal properties, are all based on a quinazolinone scaffold3,4. Patel et al. had described a series of 7-chloro-2-phenylquinazolinone derivatives as promiscuous EGFR (T790M mutation) inhibitors5.
Chalcones are unsaturated ketones, and are pharmacophoric scaffolds in many anticancer chemicals, including both synthetic and natural medicines (e.g. isoliquiritigenin, butein, etc.)6,7. Chalcones are Michael acceptors because of their unsaturated carbonyl site, which enables them to interact with a wide range of biological molecules while also tolerating them. This enables the chalcones to exhibit a variety of biological activities8.The molecular hybridization approach has been one of the most important advancements in the field of medicinal chemistry and drug discovery. In the drug design process known as molecular hybridization, two different bioactive scaffolds are linked together to form a new hybrid molecule9. The molecular hybridization strategy has been proven to be a boon for the design of novel anticancer compounds10, for example, 3-methylquinazolinone incorporated chalcone hybrids11, 4-aminoquinazoline-chalcone hybrids12, quinoline-chalcone conjugates13, chalcone-benzimidazole hybrids14, thiazole incorporated chalcone hybrids15, pyrazole-chalcone hybrids16, etc., all support using the chalcone structural motif to find new anticancer drugs. EGFR overexpression is a significant contributor in the poor prognosis and low survival rate of lung and colorectal cancer patients17,18. The driving force behind the development of EGFR -TKIs has been the poor prognosis and severe toxicities associated with conventional chemotherapeutic treatments19.
To treat NSCLC and mCRC , EGFR inhibitors of the first, second, and third generations, are currently used20. First-generation EGFR inhibitors that are 4-aminoquinazoline derivatives, are ineffective for treating NSCLC linked to the T790M EGFR mutation5,19. Afatinib and dacomitinib, which are 4-aminoquinazoline derivatives and second-generation EGFR inhibitors, proved to be effective against EGFR with the T790M mutation in preclinical models, but they failed in clinical trials because of their dose-limiting toxicity21,22. In the EGFR signaling pathways, RAS serves as a master element. One of the main causes of EGFR-targeted therapy’s failure is caused by mutations in RAS, which render the RAS protein consistently active and insensitive to upstream signals triggered by EGFR. Of these genomic alterations, KRAS mutation is the most prevalent, occurring in 40% of mCRC23. Gly12, Gly13, and Gln61 are the three locations in KRAS where mutations are most frequently seen. The most frequent mutation type is G12D and is one of the most significant targets for drug discovery as a result24.Abnormal K-RAS signaling is involved in 30% of all cases of human cancer25.K-RAS inhibition is therefore a promiscuous approach for many malignancies.
The current study concentrates on the synthesis and in vitro cytotoxicity testing of quinazolinone-chalcone hybrid ligands on normal cell lines (Vero), colorectal cancer cell lines (Caco2), and lung cancer cell lines (A549). The current research also focuses on the molecular docking of the ligands to evaluate their ability to inhibit EGFR (T790M mutation) and K-RAS (G12D mutation).As a result, the current work’s objective is to search for cytotoxic chemical entities with multi-targeted virtue that can inhibit both, mutated EGFR as well as mutated K-RAS and can exhibit selective cytotoxicity.
Materials and Methods
All the chemical reagents/solvents used were of synthetic/commercial grade. Monitoring of chemical reactions and determination of Rf values were done by TLC technique. Purification of the synthesized compounds was done by washing and recrystallization using appropriate solvents. The open capillary method was used to determine the melting point of the synthesized compounds. The structural characterization of the compounds was done by infrared FTIR-spectroscopy (Alpha-E BRUKER spectrometer), 1H-NMR ((400MHz by ECS Zeol spectrometer), and 13C-NMR spectroscopy (500MHz/400MHz by ECS Zeol spectrometer), and the molecular weight of all the synthesized compounds were confirmed by mass spectroscopy (Xevo G2-S Q Tof- Waters, USA; ESI+). In in-vitro cytotoxic studies, absorbance readings were taken using a microplate reader (Bio-Rad 550).
MarvinSketch version 18.23 (Chemaxon Ltd; http://www.Chemaxon.com) was used to create the ligand structures and their energy minimization. The target proteins were Kirsten rat sarcoma (K-RAS) with the G12D mutation (PDB ID: 4EPT) and the endothelial growth factor receptor (EGFR) with the T790M mutation of (PDB ID: 5Y9T), both of which were retrieved from the Protein Data Bank ( https://www.rcsb.org). Molecular docking investigations were conducted using AutoDock 4.0 MGL tools (The Scripps Research Institute, Molecular Graphics Laboratory, 10550 North Torrey Pines Road, CA, 92037). All docking experiments were conducted using an Intel (R) core i5 CPU running at 1.7 GHz, 3.8 GB of RAM, and the Microsoft Windows 10 operating system. The ligand-target interactions were observed using the discovery studio visualizer 2017 R2 of Dassault Systèmes BIOVIA. To calculate the RMSD value, UCSF Chimera (https://www.cgl.ucsf.edu/chimera) was used.
Chemical synthesis of 2-phenyl-4H-3,1-benzoxazin-4-one (B1),2-(4-methoxyphenyl)-4H-3,1-benzoxazin-4-one (B2),2-(4-chlorophenyl)-4H-3,1-benzoxazin-4-one (B3) and 2-(2-furyl)-4H-3,1-benzoxazin-4-one (B4)
The reaction was performed out as outlined in scheme.1(Fig.1.), dry pyridine (30 ml) was used to dissolve anthranilic acid (0.01 mol), and the resultant solution was then chilled by employing an ice bath for 20–25 minutes. Dropwise addition of aroylchloride (0.021 mol) to the ice-cold solution was carried out to keep the reaction mixture’s temperature below 10oC. In an ice bath, the reaction mixture was stirred for 15 minutes, followed by stirring at room temperature for 30 minutes to 1 hour, and then kept aside at room temperature for 15 to 40 minutes with intermittent shaking. The reaction mixture was poured into 200 ml of ice-cold water, and 10% sodium bicarbonate solution was added until the effervescence subsided. By filtration, the resulting solid precipitate was separated and repeatedly washed using ice-cold water until the filtrate became neutral to pH paper. The solid product obtained was dried and recrystallized using ethanol26,27.
Chemical synthesis of 3-(4-acetylphenyl)-2-phenylquinazolin-4(3H)-one (Q1), and 3-(4-acetylphenyl)-2-(2-furyl)quinazolin-4(3H)-one (Q4)
The fusion reactions between 4-aminoacetophenone (0.01 mol) and B1 (0.01 mol)/B4 (0.01 mol) were conducted28 as outlined in scheme.1(Fig.1.).Glacial acetic acid was used to triturate the fused mass, and the resulting suspension was added into cold water (about 200 ml). The solid obtained, was separated by filtration and repeatedly washed with water to get the filtrate neutral to pH paper, followed by methanol washing. The product thus obtained was dried and purified by recrystallization twice with methanol.
Chemical synthesis of 3-(4-acetylphenyl)-2-(4-methoxyphenyl)quinazolin-4(3H)-one(Q2) and 3-(4-acetylphenyl)-2-(4-chlorophenyl)quinazolin-4(3H)-one (Q3)
4-Aminoacetophenone (0.01 mol) and B2 (0.01 mol)/ B3 (0.01 mol) were refluxed in glacial acetic acid28 as outlined in scheme.1(Fig.1.).The reaction mixture was added to crushed ice, the solid obtained, was separated by filtration and repeatedly washed with water to get neutral pH filtrate, followed by methanol washing. The product thus obtained was dried and purified by recrystallization twice with methanol.
Chemical synthesis of 2-aryl-3-(4-((E)-3-(naphthalene-1-yl)acryloyl)phenyl)- quinazolin-4(3H)-one compounds (N1-N4):
The reactions were carried out as outlined in scheme.1(Fig.1.). Approximately 6 ml of ethanol and 3 ml of 10% NaOH (Aq.) were added to the 2-aryl-3-(4-acetylphenyl)-quinazolin-4(3H)-one compound (0.004 mol), and the resulting reaction mixture was stirred at room temperature for 10 minutes. 1-Naphthaldehyde was then added dropwise to the reaction mixture (0.004 mol)29,30. Until the reaction was completed, the reaction mixture was stirred. The reaction mixture was added to ice-cold water, and the solid precipitate obtained was separated by filtration, and repeatedly washed with water to get neutral pH filtrate. The dried product was recrystallized with acetic acid.
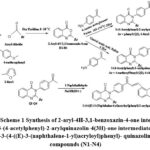 |
Figure 1: Scheme Synthesis of 2-aryl-4H-3,1-benzoxazin-4-one intermediates (B1-B4), 3-(4-acetylphenyl)-2-arylquinazolin-4(3H)-one intermediates (Q1-Q4) and 2-aryl-3-(4-((E)-3-(naphthalene-1-yl)acryloyl)phenyl)- quinazolin-4(3H)-one compounds (N1-N4). |
In-Vitro cytotoxic studies
In vitro cytotoxic studies were done by the MTT assay method31.The cell lines were obtained from NCCS, (The National Centre for Cell Science, Pune (India). The cell culture (A549, Caco2, and Vero cell lines) was centrifuged and using a DMEM medium containing 10% fetal bovine serum, the cell suspension was prepared (1.0×105 cells/ml). 100 μl of the diluted cell suspension was added to each well. After 24 hours, on attaining the adequate cell population, the centrifugation was done, and the 100μl of different test sample concentrations that had been made in the maintenance medium were added. At 37oC for 48 hrs, the plates were incubated in a 5% CO2 environment. and observations were recorded every 24 hours. 20 μl of MTT solution (concentration: 2mg/ml) in MEM-PR (MEM without phenol red) was added after 48 hours. The plates were gently shaken before being incubated at 37°C for two hours in a 5% CO2 environment. The 100 ml of DMSO was added, and the plates were gently shaken to dissolve the formazan that had formed. Using a microplate reader, the absorbance measurement was made at a wavelength of 540 nm (Bio-Rad 550).
Docking studies
Preparation of target proteins
The procedure is comprised of several steps32,33. First, the crystal structures of the naquotinib complexed mutated EGFR (T790M mutation; PDB Id: 5Y9T), mutated human K-RAS (G12D mutation; Pdb Id: 4EPT) complexed with 2-hydroxyphenyl pyrrolidin-1-yl and GDP, in .pdb file type, were downloaded from protein data bank . The complexed ligand-protein crystal structures were opened in discovery Studio Visualizer to separate the co-crystallized ligands and water molecules. Using the Auto Dock 4.0 MGL tools, the revised 3D crystal structures of the target proteins were prepared for docking experiments by adding the polar hydrogen atoms, the merging of non-polar hydrogens, and the inclusion of Kollman charges, and were then saved in the .pdbqt file format.
Ligand preparation
The 2D structures were drawn using MarvinSketch, and subsequently converted to 3D structures. Explicit hydrogens were added to each ligand, the geometry of the ligands was then cleaned, and the ligands’ energy was minimized in an MMFF94 force field using Marvin Sketch’s gradient optimization function. The ligands’ 3D structures were stored as .pdb files. The autodock tools program was used to open the ligand structure (.pdb file format). After giving all of the ligand’s atoms the AD4 type, merging non-polar hydrogen atoms, adding polar hydrogen atoms, and inclusion of gasteiger charges, every ligand structure was stored in the .pdbqt file format, for docking experiments 32,33.
Docking run
For target 5Y9T
For autogrid computing and docking investigations, respectively, Autogrid-4 and Autodock-4 programs of MGL tools were employed33.To create the grid parameter file (.gpf format) for each ligand, a 3D grid box was put in the centre of the target macromolecule, and 90 grid points were selected along its x, y, and z axis with a grid spacing of 0.375 A °.For each ligand, a grid log file (.glg file format) was created using Autogrid-4 of the MGL tools from its respective grid parameter file (.gpf format). In the preparation of the docking parameter file (.dpf file format), a genetic algorithm technique was employed, docking experiment was set for 100 independent runs such that each run contained an initial population of 150 conformations. For picking the worst individual, the number of generations was set to 10, and an elitism value of 1 was set for cluster analysis. Autodock4.0 was run to start docking with the .dpf file format of the ligand that generated a docking log file (.dlg file format) of the ligand under study.
For target 4EPT
All the process adopted was the same as aforementioned for 5Y9T, but for this target 100 grid points were selected.
Docking data, observation of docked conformation and interactions with macromolecule
Ligand’s free energy of binding (BE) and inhibition constant (KI) were obtained from their respective .dlg files. The complex between the ligand’s best fit conformer and its respective target was saved in .pdb file format. To visualize the ligand’s binding pose and its interactions with the amino acid residues of the target macromolecule, the .pdb file of the complex was opened and viewed in Discovery Studio Visualizer.
Results and discussion
Chemical synthesis
Compound B1
Molecular formula:C14H9NO2 ; Molecular weight: 223.22676 ; White solid; Melting point: 120-122oC; Rf ( 30% ethylacetate + 70% hexane): 0.674; Reaction time: 1hour 10 minutes; Yield: 73.58% . Mass Spectra (ESI+): Calculated for [C14H9NO2+H+]: 224.22676, found 224.2261; IR-ATR (ν, cm-1): 1759.21 (lactone C=O str), 1568(C=N str); 1H- NMR (400 MHz, CDCl3) δ ppm: 8.31-7.5 (m, 9H, Ar-H); 13C- NMR (400 MHz CDCl3) δ ppm: 159.66 (1C, C=O of benzoxazin-4-one), 157.12(1C, N=C-O of benzoxazin-4-one), 147.06 (1C, Aromatic), 136.65 (1C, Aromatic),132.71(1C, Aromatic), 130.31(1C, Aromatic), 128.83(2C, Aromatic), 128.68(1C, Aromatic), 128.4 (2C, Aromatic), 128.35 (1C, Aromatic), 127.32(1C, Aromatic), 117.1(1C, Aromatic)
Compound B2
Molecular formula: C15H11NO3;Molecular weight: 253.25274;White solid; Melting point: 142-145oC ; Rf ( 30% ethylacetate + 70% hexane): 0.628; Reaction time: 1 hour 55 minutes; Yield: 67.1%.MS(ESI+):Calculated for [C15H11NO3+H+]: 254.25274, found 254.2527; IR-ATR (ν, cm-1): 1753.48 ((lactone C=O str), 1593.25(C=N str); 1H- NMR (400 MHz,CDCl3) δ ppm: 8.24-6.95(m,8H,Ar-H), 3.86 (s,3H, CH3 of methoxy) ; 13C-NMR (500MHz, DMSO-d6) δ ppm: 163.38 (1C,Aromatic), 159.51(1C, C=O of benzoxazin-4-one), 156.89 (1C, N=C-O of benzoxazin-4-one),147.08 (1C, Aromatic), 137.34(1C, Aromatic), 130.32(2C, Aromatic), 128.57(1C, Aromatic), 128.53 (1C, Aromatic), 127.12(1C, Aromatic), 122.58 (1C, Aromatic), 117.05(1C, Aromatic), 114.96(2C, Aromatic), 56.06 (1C, CH3 of methoxy)
Compound B3
Molecular formula: C14H8ClNO2; Molecular weight:257.67182;White solid; Melting point: 183- 185oC ; Rf ( 30% ethylacetate + 70% hexane): 0.744; Reaction time: 1 hour; Yield: 75.03% .MS(ESI+): Calculated for [C14H8ClNO2+H+]: 258.67182, found 258.6718; IR-ATR (ν, cm-1): 1766.43 (lactone C=O str), 1620.60 (C=N str); 1H- NMR (400 MHz,CDCl3) δ ppm:8.23-7.22(m,8H,Ar-H); 13C-NMR(500MHz CDCl3) δ ppm: 159.3(1C, C=O of benzoxazin-4-one), 156.24 (1C, N=C-O of benzoxazin-4-one),146.8(1C, Aromatic), 139.08(1C, Aromatic),136.69(1C, Aromatic),129.62(2C, Aromatic), 129.13(2C, Aromatic),129.01(1C, Aromatic), 128.74(1C, Aromatic),128.69(1C, Aromatic), 127.26(1C, Aromatic), 116.97(1C, Aromatic)
Compound B4
Molecular formula: C12H7NO3;Molecular weight:213.18888;White solid; Melting point: 102- 105oC; Rf ( 30% ethylacetate + 70% hexane): 0.593; Reaction time: 1hour 5 minutes; Yield: 65.6%. MS(ESI+):Calculated for [C12H7NO3+H+]: 214.18888, found 214.1889; IR-ATR (ν, cm-1):1750.99 (lactone C=O str),1638.48 (C=N str); 1H NMR (400 MHz, CDCl3) δ ppm: 8.19-6.58 (m,7H, Ar-H and Furan-H);13CNMR (500MHz,DMSO-d6) δ ppm: 163.32 (1C, N=C-O of benzoxazin-4-one),158.76 (1C, C=O of benzoxazin-4-one),148.38 (1C, Aromatic), 146.74 (1C, Aromatic), 144.56 (1C, Aromatic), 137.4 (1C, Aromatic), 128.86 (1C, Aromatic), 128.66(1C, Aromatic), 127.13(1C, Aromatic), 117.52(1C, Aromatic), 117.44(1C, Aromatic), 113.46 (1C, Aromatic)
Compound Q1
Molecular formula: C22H16N2O2; Molecular weight: 340.37464; Light yellow solid; Melting point: 165-168oC ; Rf ( 30% ethylacetate + 70% hexane):0.354;Yield: 51.57%. MS(ESI+):Calculated for [C22H16N2O2+H+]: 341.37464, found 341.3746; IR-ATR (ν, cm-1): 1679.45 & 1649.75 (C=Os str), 1586.59 (C=N str); 1H-NMR (400 MHz, DMSO-d6) δ ppm: 8.28 (d,1H, Ar-H), 7.91-7.22 (m,12H, Ar-H), 2.47 (s,3H, CH3); 13CNMR (500MHz,DMSO-d6) δ ppm: 197.16 (1C, C=O of acetyl), 168.10 (1C, N=C-N of quinazolin-4-one ring), 165.27 (1C, C=O of quinazolin-4-one ring),143.61(1C, Aromatic),138.73(1C, Aromatic), 134.92(1C, Aromatic),132.80(1C, Aromatic), 132.51 (1C, Aromatic), 129.73(2C, Aromatic), 129.6(1C,Aromatic), 129.61(2C, Aromatic), 129.3(2C, Aromatic), 127.60(1C, Aromatic), 127.49(1C, Aromatic), 124.04(1C, Aromatic), 122.39(1C, Aromatic),120.47 (2C, Aromatic), 26.97 (1C, CH3 of acetyl).
Compound Q4
Molecular formula C20H14N2O3; Molecular weight: 330.33676;Brown solid; Melting point: 180-182oC; Rf ( 30% ethylacetate + 70% hexane): 0.316;Yield: 53.03%. MS(ESI+): Calculated for [C20H14N2O3+H+]: 330.33676, found 331.3368; IR-ATR (ν, cm-1): 1672.70 & 1651.30 (C=Os str),1582.70 (C=N str); 1H-NMR (400 MHz, DMSO-d6) δ ppm: 8.36(d, 1H, Ar-H), 7.93-6.65 (m,10H,Ar-H & Furan-H), 2.59 (s, 3H, CH3); 13C-NMR (400MHz,DMSO-d6) δ ppm:197.18 (1C, C=O of acetyl), 168.08 (1C, N=C-N of quinazolin-4-one ring), 164.24(1C, C=O of quinazolin-4-one ring), 147.83(1C, Aromatic), 146.64(1C, Aromatic), 143.5(1C, Aromatic), 138.45(1C, Aromatic), 133.03(1C, Aromatic), 132.89(1C, Aromatic), 129.76(2C, Aromatic), 129.69 (1C, Aromatic),126.97(1C, Aromatic),123.85(1C, Aromatic), 121.74(1C, Aromatic), 120.58(2C, Aromatic), 115.94(1C, Aromatic), 113.14(1C, Aromatic), 27.02 (1C,CH3 of acetyl).
Compound Q2
Molecular formula: C23H18N2O3; Molecular weight: 370.40062; White solid; Melting point: 185-190oC ; Rf ( 30% ethylacetate + 70% hexane):0.266; Yield: 52.9%; MS(ESI+): Calculated for [C23H18N2O3+H+]: 371.40062, found 371.4007; IR-ATR (ν, cm-1): 1672.64 & 1643.96 (C=Os str), 1580.83 (C=N str); 1H NMR (400 MHz,DMSO-d6) δ ppm: 8.31(d,1H, Ar-H),7.97-7.05(m,11H,Ar-H), 3.78 (s, 3H, CH3 of methoxy), 2.48(s, 3H, CH3 of acetyl); 13CNMR (400MHz,DMSO-d6) δ ppm: 197.15(1C, C=O of acetyl), 168.24 (1C,N=C-N of quinazolin-4-one ring), 164.68(1C, C=O of quinazolin-4-one ring),162.8 (1C, Aromatic), 143.6 (1C, Aromatic),139.12(1C, Aromatic), 134.83 (1C, Aromatic) 132.81(1C, Aromatic), 129.74(1C, Aromatic), 129.52(2C, Aromatic),129.45(2C, Aromatic),127.06(1C, Aromatic), 123.67(1C, Aromatic), 123.12(1C, Aromatic), 120.5(2C, Aromatic), 120.2(1C, Aromatic), 114.73(2C, Aromatic), 56 (1C, CH3 of methoxy), 27 (1C,CH3 of acetyl).
Compound Q3
Molecular formula C22H15ClN2O2; Molecular weight:374.8197; White solid; Melting point:190-192oC; Rf ( 30% ethylacetate + 70% hexane): 0.380; Yield:54.6%; MS(ESI+):Calculated for [C22H15ClN2O2+H+]:375.8197, found 375.8196; IR-ATR (ν, cm-1): 1665.24 (C=Os str),1586.56 (C=N); 1H NMR (400 MHz, DMSO-d6) δ ppm: 8.56 (d,1H,Ar-H), 7.88-7.11(m,11H, Ar-H), 2.41(s,3H, CH3); 13CNMR (400MHz,DMSO-d6) δ ppm : 197.12(1C,C=O of acetyl), 167.94 (1C, N=C-N of quinazolin-4-one ring),164.34(1C, C=O of quinazolin-4-one ring),143.69 (1C, Aromatic), 138.33(1C, Aromatic), 137.52(1C, Aromatic),133.78(1C, Aromatic),132.74(1C, Aromatic), 130.27(1C, Aromatic),129.74(2C, Aromatic),129.59 (2C, Aromatic),129.43(2C, Aromatic) 129.4(2C, Aromatic), 127.41(1C, Aromatic), 123.67(1C, Aromatic), 120.51(2C, Aromatic), 26.99(1C, CH3 of acetyl).
Compound N1
Molecular formula C33H22N2O2; Molecular weight:478.53998 ; Yellow solid; Melting point: 220-222oC; Reaction time: 3hours; Rf ( 30% ethylacetate + 70% hexane): 0.500;Yield: 61.12%. MS(ESI+): Calculated for [C33H22N2O2+H+]: 479.53998, found 479.5390; IR-ATR (ν, cm-1): 1644.49 (C=Os str), 1593.77 (C=N str);1H- NMR (400 MHz,CDCl3) δ ppm: 8.69 (d,1H,ethylenic), 8.53(d,1H,Ar-H),8.26 (d,1H,Ar-H), 8.17 (d,2H,Ar-H), 8.00 (d,2H,Ar-H),7.95 (d, 2H,Ar-H), 7.86(d,1H,Ar-H),7.90(d,1H,Ar-H),7.65 (d,1H,ethylenic),7.61-7.49(m,8H, Ar-H), 7.34 (t,1H,Ar-H), 6.91(t,1H,Ar-H); 13C-NMR(400MHz,CDCl3) δ ppm: 188.54 (1C, C=O of chalcone structural motif), 168.39 (1C, N-C=N of quinazolinone ring), 165.29 (1C, C=O of quinazolinone ring), 142.97(1C, Aromatic),140.9 (1C, C=C site of chalcone structural motif), 139.78(1C,Aromatic), 134.54(1C, Aromatic), 133.57(1C, Aromatic), 132.78(1C, Aromatic), 132.13(1C, Aromatic), 131.87(1C, Aromatic), 131.55(1C, Aromatic), 130.66(1C, Aromatic),129.58(2C,Aromatic),129.59(2C, Aromatic), 128.8(1C, Aromatic), 128.7(3C, Aromatic), 127.17 (2C, Aromatic), 126.89(1C, Aromatic), 126.22(1C, Aromatic), 125.45(1C, Aromatic), 125.03(1C, Aromatic), 124.33(1C, Aromatic), 123.25(1C, Aromatic), 122.61(1C, Aromatic), 121.3(1C, C=C site of chalcone structural motif),120.45(2C, Ar).
Compound N2
Molecular formula: C34H24N2O3; Molecular weight:508.56596 ; Yellow solid; Melting point: 225-228oC; Reaction time: 3hours; Rf ( 30% ethylacetate + 70% hexane): 0.450;Yield: 46%.MS(ESI+):Calculated for [C34H24N2O3+H+]: 509.56596, found 509.5650; IR-ATR (ν, cm-1): 1646.97 (C=Os str), 1569.27(C=N str ); 1H NMR (400 MHz,CDCl3) δ ppm:8.69 (d,1H,ethylenic), 8.36 (d, 1H,Ar-H), 8.26 (d,1H,Ar-H), 8.17(d,2H,Ar-H), 8.02 (d, 2H,Ar-H),7.96 (d, 2H,Ar-H),7.92(d, 1H,Ar-H), 7.88 (d, 1H,Ar-H), 7.67 (d,1H,ethylenic), 7.58( t,1H,Ar-H),7.55-7.47 (m,4H,Ar-H),7.19 (t,1H,Ar-H), 7.15 (d,2H,Ar-H), 6.75 (t,1H,Ar-H), 3.88( s,3H,OCH3); 13C-NMR(400MHz,CDCl3) δ ppm: 188.93(1C,C=O of chalcone structural motif), 167.88 (1C, N=C-N of quinazolinone ring), 165.88(1C, C=O of quinazolinone ring), 162.92 (1C, Aromatic), 142.52(1C, Aromatic), 141.63(1C, C=C of chalcone structural motif), 139.04(1C,Aromatic),134.16(1C, Aromatic), 133.8(1C, Aromatic), 132.37(1C, Aromatic), 131.84(1C, Aromatic), 131.51(1C,Aromatic) 130.88(1C, Aromatic), 130.18(2C, Aromatic), 129.41(2C, Aromatic), 129.4(1C, Aromatic),128.84(1C, Aromatic), 127.08(1C, Aromatic), 126.57(1C, Aromatic), 126.4(1C, Aromatic), 125.53(1C, Aromatic), 125.16(1C, Aromatic), 124.47(1C, Aromatic), 123.6(1C, Aromatic), 122.97(1C, Aromatic),122.07(1C, Aromatic), 121.72(1C, C=C of chalcone structural motif), 119.92(2C, Aromatic), 114.23(2C, Aromatic), 55.58(1C, methyl of -OCH3)
Compound N3
Molecular formula: C33H21ClN2O2; Molecular weight:512.98504 ;Light yellow solid; Melting point: 227-229oC; Reaction time: 3hours; Rf ( 30% ethylacetate + 70% hexane): 0.600; Yield: 52.2%. MS(ESI+): Calculated for [C33H21ClN2O2+H+]: 513.98504, found 513.9857; IR-ATR (ν, cm-1): 1650.47(C=Os str),1583.77 (C=N str); 1H- NMR (400 MHz,CDCl3) δ ppm: 8.66(d,1H,ethylenic), 8.56(d,1H,Ar-H), 8.23(d,1H,Ar-H), 8.14(d, 2H,Ar-H),7.92( d, 2H,Ar-H),7.88(d,2H,Ar-H),7.86 (d, 1H,Ar-H), 7.8(d,1H,Ar-H), 7.62 (d,1H,ethylenic), 7.58-7.50(m, 5H, Ar-H),7.47( d, 2H,Ar-H),7.38( t, 1H,Ar-H), 6.97(t, 1H,Ar-H). 13C-NMR(400MHz, DMSO-d6) δ ppm: 188.06(1C, C=O of chalcone structural motif), 168(1C,N=C-N of quinazolinone ring), 164.34(1C,C=O of quinazolinone ring), 143.93(1C, Aromatic), 141.68(1C, C=C of chalcone structural motif), 138.37(1C, Aromatic), 137.29 (1C, Aromatic),133.86 (1C, Aromatic),133.74(1C, Aromatic),132.69(1C, Aromatic), 131.51(1C,Aromatic),131.89(1C, Aromatic), 130.74(1C,Aromatic), 130.26(2C, Aromatic),130.18(1C,Aromatic), 129.61(3C, Aromatic), 129.39(2C, Aromatic), 129.29(1C,Aromatic), 127.75(1C, Aromatic), 126.81(1C, Aromatic), 126.2(1C, Aromatic), 125.15(1C, Aromatic), 124.97(1C, Aromatic), 124.33(1C, Aromatic), 123.49(1C, Aromatic), 122.82(1C, C=C of chalcone structural motif), 120.56 (2C, Aromatic), 120.39(1C, Aromatic).
Compound N4
Molecular formula:C31H20N2O3; Molecular weight:468.5021;Yellow solid; Melting point: 218-220oC; Reaction time: 3hours; Rf ( 30% ethylacetate + 70% hexane): 0.475; Yield: 50.1%. MS(ESI+):Calculated for [C31H20N2O3+H+]: 469.5021, found 469.5020; IR-ATR (ν, cm-1):1648.01(C=Os str ), 1581.49 (C=N str);1H-NMR ( CDCl3 400MHz) δ ppm: 8.68 (d,1H,ethylenic), 8.48( d,1H,Ar-H),8.25 (d,1H,Ar-H), 8.14 (d,2H,Ar-H),7.91(d,2H,Ar-H),7.88(d,1H,Ar-H),7.75 (d,1H,Ar-H),7.64 (d,1H,ethylenic), 7.6-7.5 ( m, 6H, Ar-H, Furan-H),7.35 (t,1H,Ar-H), 7.24 (d,1H,Furan-H), 6.93 (t, 1H,Ar-H), 6.54 ( dd, 1H,Furan-H); 13C-NMR(400MHz,CDCl3) δ ppm: 188.92(1C, C=O of chalcone structural motif),167.46(1C, N=C-N of quinazolinone ring), 164.44(1C, C=O of quinazolinone ring), 147.87(1C, Aromatic), 145.36(1C, Aromatic), 142.1(1C, Aromatic), 141.74(1C, C=C of chalcone structural motif),138.67(1C,Aromatic), 133.8(1C, Aromatic), 132.91(1C, Aromatic), 132.47(1C, Aromatic), 131.84(1C, Aromatic), 131.53(1C, Aromatic),130.92(1C, Aromatic),130.18(2C, Aromatic), 128.85(1C, Aromatic), 127.36(1C, Aromatic), 127.09(1C, Aromatic), 126.41(1C, Aromatic), 125.53(1C, Aromatic), 125.18(1C, Aromatic),124.41(1C, Aromatic), 123.58(1C, Aromatic), 123.34(1C, Aromatic), 122.13(1C, C=C of chalcone structural motif), 121.62(1C, Aromatic), 120.01(2C, Aromatic), 115.71(1C, Aromatic), 112.42(1C, Aromatic).
Cytotoxic studies
The in-vitro cytotoxic potential (by MTT method) of the title compounds is summarized in Table 1.
Table 1: Cytotoxicity (IC50 μg/ml ) of the compounds(N1-N4)
|
Compound |
Name of the cell line (IC50 values μg/ml) |
||
|
A549 |
Caco2 |
Vero |
|
|
N1 |
12.38 |
22.69 |
15.02 |
|
N2 |
15.50 |
15.95 |
76.62 |
|
N3 |
11.29 |
10.79 |
18.72 |
|
N4 |
19.87 |
15.81 |
28.86 |
|
Erlotinib |
44.41 |
16.13 |
25.78 |
In-vitro cytotoxicity for the A549 cell lines
The in-vitro investigations showed that all of the title compounds (N1-N4) are more cytotoxic than the reference erlotinib (IC50: 44.41 μg/ml), with values ranging from 11.29 to 19.87 (μg/ml). The most potent cytotoxic compound against lung cancer cell lines A549 is N3 (IC50: 11.29 μg/ml), followed by N1 (IC50: 12.38 μg/ml).
In-vitro cytotoxicity for the Caco2 cell lines
The in-vitro investigations showed that, except for N1 (IC50: 22.69 μg/ml), the other title compounds (N2-N4) are more cytotoxic than the reference erlotinib (IC50: 16.13 μg/ml), with IC50 (μg/ml) values ranging from 10.79 to 15.95. The cytotoxicity of compounds N2 (IC50: 15.95 μg/ml) and N4 (IC50: 15.81 μg/ml) is very similar to that of the reference drug erlotinib.
In-vitro cytotoxicity for the normal cell lines (Vero)
The in-vitro investigations showed that the compounds N2 and N4 are less toxic to normal cells than the reference erlotinib (IC50:25.78 μg/ml), with IC50 values of 76.62 μg/ml and 28.86 μg/ml, respectively. The compound N2, which is least toxic to normal cells, has a high selectivity for lung and colorectal cancer cells.
Molecular docking studies
Docking Analysis (with 5Y9T)
Every docked ligand binded to the 5Y9T target’s reported active site34, as shown in Fig. 2.and Fig.3., ensuring the good affinity of the ligands under study and, the accuracy of the docking experiments. Docking analysis as summarized in Table 2, revealed that all the ligands have good affinity for the active site of the target protein (5Y9T) as they have high negative binding energies (ranging from -45.27 to -43.14 KJ/mol), and the inhibition constant in nanomoles (ranging from 11.63 to 34.69 nmol).
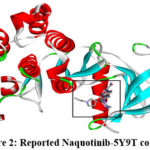 |
Figure 2: Reported Naquotinib-5Y9T complex |
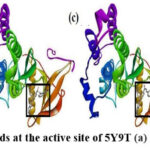 |
Figure 3: Docked ligands at the active site of 5Y9T (a) N1 (b) N2 (c) N3 (d) N4 |
Table 2: Docking analysis (with 5Y9T)
|
Target protein 5Y9T |
||||||
|
Compound |
Binding Energy(B.E.) (KJ/mol) |
Inhibition Constant (Ki) (nM) |
No. of H-bonds |
H-bond interaction residues (Bond distance in Ao) |
Other Interaction residues (Polar and non polar) |
No. of interacting residues |
|
N1 |
-44.18 |
18.33 |
1 |
MET793 (2.38) |
LEU792,LYS728,PRO794, |
8 |
|
N2 |
-43.14 |
34.69 |
1 |
MET793 (2.29) |
LEU792,LYS728,PRO794,LEU718, |
12 |
|
N3 |
-43.26
|
26.32
|
1
|
MET793(2.12)
|
PHE723,ALA743, MET790,ASP855,LYS745,LYS728, |
11
|
|
N4 |
-45.27 |
11.63 |
2 |
MET793(1.93),LYS 745(1.94) |
ALA743,LEU844,ALA722,VAL726,LYS728, |
11
|
Interaction of N1 with 5Y9T
As shown in Fig.4, the α-β unsaturated carbonyl group’s oxygen atom in the N1 compound is involved in H-bonding with MET793 residue, phenyl substituent at 2nd position of quinazolinone scaffold is in π -anion polar interaction with ASP855 residue, naphthalene ring system is the site for π -lone pair and π -sigma interactions with PRO794 and LEU718, respectively. All aromatic sites of the compound are involved in π-alkyl interactions and, the amino acid residues involved are LEU792, LYS728, PRO794, LEU718, VAL726, and LEU844.
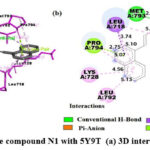 |
Figure 4: Interaction of the compound N1 with 5Y9T (a) 3D interactions (b) 2D interactions |
π
Interaction of N2 with 5Y9T
As shown in Fig.5, the α-β unsaturated carbonyl group’s oxygen atom in the N2 compound is involved in H-bonding with MET793 residue, the phenyl ring at the 2nd position of quinazolinone scaffold is the site for π -anion polar interaction with ASP855 residue, naphthalene ring system makes π -lone pair and π -sigma interactions with PRO794 and LEU718, respectively. Benzene ring of quinazolinone makes π – π interaction with PHE723.The naphthalene ring system, phenyl ring at the 3rd position of quinazolinone scaffold, and methyl of methoxy substituent involved in π – alkyl interactions and, amino acid residues involved are LEU792, LYS728, PRO794, LEU718, ALA743, LEU844, and VAL726.
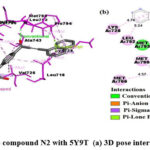 |
Figure 5: Interaction of the compound N2 with 5Y9T (a) 3D pose interactions (b) 2D interactions |
Interaction of N3 with 5Y9T
As shown in Fig.6, in the N3 compound, the α-β unsaturated carbonyl group’s oxygen atom is involved in H-bonding with MET793 residue, the 4-chlorophenyl substituent at the 2nd position of quinazolinone is the site for π -cation and π -anion interactions with LYS745 and ASP855, respectively. The naphthalene ring system makes π -lone pair interaction with PRO794 and π -sigma interaction with LEU718 and LYS728. Benzene ring of quinazolinone makes π – π interaction with PHE723. The aromatic rings of the molecule are involved in π -alkyl interactions and the residues involved are LEU792, LYS728, PRO794, LEU718, ALA743, LEU844, and VAL726.
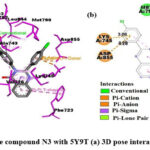 |
Figure 6: Interaction of the compound N3 with 5Y9T (a) 3D pose interactions (b) 2D interactions |
Interaction of N4 with 5Y9T
As shown in Fig.7, In theN4 compound, the oxygen atom of the α-β unsaturated carbonyl group forms H-bond with MET793, while the furan ring oxygen forms a hydrogen bond with LYS745. The furan ring is also involved in π -anion interaction with ASP855. The phenyl ring at the 3rd position of quinazolinone is involved in π -sulphur interaction with MET790. The naphthalene ring system makes π -lone pair interaction with PRO794 and π -sigma interaction with LEU718. Benzene ring of quinazolinone makes π – π interaction with PHE723. All the aromatic ring systems except furan are involved in π -alkyl interactions and the residues involved are LEU844, LEU792, ALA743, LEU718, and VAL726.
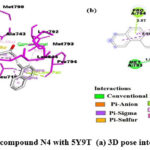 |
Figure 7: Interaction of the compound N4 with 5Y9T (a) 3D pose interactions (b) 2D interactions |
Docking analysis (with 4EPT)
Every docked ligand binds to the macromolecule’s stated GDP binding site25 of the target protein (4EPT) as shown in Fig.8 and Fig.9., ensuring the good affinity of the ligands for the active site of the target protein and, the accuracy of the docking experiments. Docking analysis with 4EPT as summarized in Table 3, revealed that all the ligands have good affinity for the active site of the target protein (4EPT) as they have high negative binding energies (ranging from -44.35 to -49.96 KJ/mol), and the inhibition constant in nanomoles (ranging from 1.78 to 16.86 nmol).
 |
Figure 8: Reported 4EPT – GDP complex |
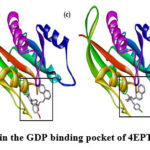 |
Figure 9: Docked ligands in the GDP binding pocket of 4EPT (a) N1 (b) N2 (c) N3 (d) N4 |
Table 3: Docking analysis (with 4EPT).
|
Target protein 4EPT |
||||||
|
Compound |
Binding energy (BE) in kJ/mol |
Inhibition constant (KI) in nmol |
Number of H bonds |
H bond interaction (A0) |
Other interacting amino acids(polar and non polar) |
No. of interacting amino acids |
|
N1 |
-49.96 |
1.78 |
3 |
SER17(2.27),ALA18(2.74)Glu31(1.77) |
LYS16,ALA59,GLY13,PRO34,ASP33, |
13
|
|
N2 |
-49.20 |
2.40 |
3 |
ALA18(2.64),SER17(2.13),GLU31(1.80) |
LYS16,ALA59, PRO34,ALA146,LYS117,
|
11 |
|
N3 |
-46.94 |
5.98 |
2 |
LYS16(2.82),TYR32(2.62) |
ALA59,PRO34,LYS117,ALA146, |
9
|
|
N4 |
-44.35 |
16.86 |
2 |
TYR32(2.21),GLU31(2.41) |
LYS16,ALA146,LYS147,ALA18,
|
10
|
Interaction of N1 with 4EPT
As shown in Fig.10, in the N1 compound, the carbonyl’s oxygen atom of the quinazolinone forms H-bonds with SER17 and ALA18, the α-β unsaturated carbonyl’s oxygen atom is also involved in H-bonding with GLU31, the benzene ring of quinazolinone is involved in π -cation interaction and, π -alkyl interaction with LYS16 and ALA59 respectively, the phenyl substituent at the 2nd position of quinazolinone ring forms π – sigma and, π -amide stacked interaction with PRO34 and ASP33 respectively. The naphthalene ring system forms π – π bonding with PHE28 and, is involved majorly in π -alkyl interactions with LYS117, LEU120, ALA146, and LYS147 respectively.
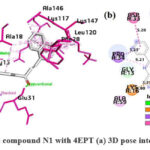 |
Figure 10: Interaction of the compound N1 with 4EPT (a) 3D pose interactions (b) 2D interactions |
Interaction of N2 with 4EPT
As shown in Fig.11, in the N2 compound, the carbonyl’s oxygen atom of the quinazolinone ring forms H-bonds with SER17 and ALA18, the benzene ring of quinazolinone is involved in π -cation interaction with LYS16, the 4-methoxyphenyl substituent at the 2nd position of quinazolinone forms π -alkyl bonding with PRO34, the α-β unsaturated carbonyl’s oxygen atom is also involved in H-bonding with GLU31, naphthalene ring system forms a π – π interaction with PHE28 and π -alkyl interactions with ALA146, LYS117, LYS147, LEU120, and ALA18.
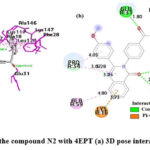 |
Figure 11: Interaction of the compound N2 with 4EPT (a) 3D pose interactions (b) 2D interactions |
Interaction of N3 with 4EPT
As shown in Fig.12, in the N3 compound, the carbonyl oxygen of the quinazolinone ring forms a H-bond with LYS16, the oxygen atom of the α-β unsaturated carbonyl group is also involved in H-bonding with TYR32, the naphthalene ring system participates in a π -cation interaction, and in a π – π interaction with LYS117 and PHE28, respectively. All the aromatic rings participate in π -alkyl bonding and the residues involved are ALA59, PRO34, ALA18, ALA146, and LYS147.
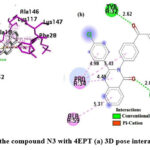 |
Figure 12: Interaction of the compound N3 with 4EPT (a) 3D pose interactions (b) 2D interactions |
Interaction of N4 with 4EPT
As shown in Fig.13, in the N4 compound, the oxygen atom of α-β unsaturated carbonyl group forms H-bond with GLU31 and TYR32, the furan ring at the 2nd position of the quinazolinone forms a π -cation interaction with LYS16, the benzene ring of quinazolinone system participates in π -alkyl interaction with PRO34, naphthalene ring system is involved in π -cation interaction and π – π interaction with LYS117 and PHE28, respectively. The naphthalene ring system is also involved in π -alkyl bonding with ALA18, LYS117, ALA146, and LYS147.
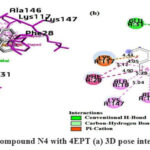 |
Figure 13: Interaction of the compound N4 with 4EPT (a) 3D pose interactions (b) 2D interactions |
The literature survey revealed that need to treat NSCLC with mutant EGFR and, to treat colorectal(CRC) with mutant KRAS is still a substantive problem that is unmet to date. The 3-methyl quinazolinone chalcone hybrids have been characterized by Wani et al. as potent anticancer agents against pancreatic cancer cell lines with IC50 values ranging from 5.5 to 8.5 μM11. Madhavi et al. have described certain 4-aminoquinazoline incorporated chalcones as potent cytotoxic agents against cancer cell lines (melanoma cancer, breast cancer, colorectal cancer, and lung cancer), with an IC50 range from 0.10 to 0.19 μM12. The antimicrotubule action (anticancer) of synthetic naphthyl chalcones has been the subject of numerous reports8.With IC50 ( μM) values ranging from 0.01-0.54, the 3-methyl-quinazolinone derivatives were reported by Le et al. as strong inhibitors of wild-type EGFR35. With IC50 values 0.047 to 2.71( μM), Zhang et al. reported certain 2-phenoxymethylquinazolinone derivatives as effective EGFR (wild- type) inhibitors36. For treating NSCLC patients with T790M-mediated resistance, the third-generation EGFR inhibitors (such as osimertinib and olmutinib) offered a potential therapeutic approach, but they are also linked to serious side effects37. For NSCLC patients who have developed resistance to EGFR TKIs because of the mutation T790M, there are still few effective therapy alternatives available37. Zhao et al. have reported certain quinazoline derivatives as KRAS-G12C inhibitors as promiscuous anticancer agents38.The rate-determining step in K-Ras activation is the transition from GDP bound K-Ras to GTP bound K-Ras. Targeting the inactive, GDP-bound KRAS (G12C) has been a promiscuous technique for creating novel anti-KRAS therapies, as seen by the FDA’s clearance of AMG 510 (Lumakras) 24,39. In the present study, the title ligands were docked to K-RAS (G12D mutation), trapping it in an inactive conformation and preventing it from coupling with the downstream effector pathway. The present work is an effort to explore a few low molecular weight chemical entities for their cytotoxic potential and, that can act smartly by inhibiting the mutant molecular targets (EGFR and K-RAS), moreover, these chemical entities can be used as a template for the development of future smart dual targeting drugs, to overcome resistance hindrance.
Conclusion
The intended chalcone hybrid compounds with quinazolinone incorporation were made in three steps. Benzoxazine-4-one intermediates (B1-B4) were synthesized in the first step, and in the second step, the intermediate compounds (B1-B4) were treated with 4-aminoacetophenone to afford intermediates with quinazolin-4-one ring (Q1-Q4). The quinazolin-4-one (Q1-Q4) intermediates and 1-naphthaldehyde underwent an aldol condensation reaction in the third step, resulting in the formation of hybrid molecules (N1-N4). The TLC method, which uses a solvent system of 30% ethyl acetate and 70% hexane, was successful in monitoring all of the chemical reactions used in the current study to produce the target hybrid compounds. Using IR, 1H-NMR, 13C-NMR, and mass spectroscopy (ESI+), the title compounds and the intermediate products were satisfactorily characterized. The results of the in-vitro cytotoxic screening and docking analysis can be concluded as follows:
(a) When used with the chalcone structural motif, quinazolin-4-one scaffolds with the aryl rings in their second and third positions function as potential chemical templates for anticancer activity.
(b) Docking experiments showed that all the synthesized quinazolinone-chalcone hybrid ligands had a strong affinity for the active site and could inhibit the EGFR (with T790M mutation; pdb Id: 5Y9T) and K-RAS (with G12D mutation; pdb Id: 4EPT).
(c) In-vitro cytotoxic screening against lung cancer cell lines (A549) and colorectal cancer cell lines revealed that N3 is the most potent compound, but N2 is the most selective towards cancer cells.
The studied title compounds can be used as a template for chemical tailoring/chemical modification to make more selective and smart cytotoxic agents with the virtue of the capability to inhibit mutant EGFR and mutant K-RAS. Therefore, the current work can be a catalyst for the development of novel low molecular weight therapeutic molecules to treat colorectal cancer and to treat lung cancer linked to EGFR and K-RAS mutation.
Acknowledgments
The MNIT- Materials research centre in Jaipur, India provided the spectral data, which the authors are grateful for.The Principal, J.S.S. College of Pharmacy, Ooty (Tamil Nadu, India), for providing the necessary laboratory facilities for invitro cytotoxicity investigations, as well as Cadila Healthcare Ltd. (Ahemdabad, India), for supplying the gift sample of erlotinib, are both acknowledged by the authors.
Conflict of interest
It is stated by the authors that they have no conflict of interests.
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018, 68,394-424. doi: 10.3322/caac.21492.
CrossRef - Rebecca, L.S.; Kimberly, D.M.; Hannah, E.F.; Ahmedin, J. Cancer statistics 2022. CA Cancer J Clin. 2022, 72,7–33. Doi 10.3322/caac.21708.
CrossRef - Hameed, A.; Al-Rashida, M.; Uroos, M.; Ali, S.A.; Arshia, I. M.; Khan, K.M. Quinazoline and quinazolinone as important medicinal scaffolds: a comparative patent review (2011-2016). Expert Opin Ther Pat. 2018, 28, 281-297. doi: 10.1080/13543776.2018.1432596.
CrossRef - Rashmi, T.; Priya, M.G.R.; Murugan, V. Quinazolinone – A Biologically Active Scaffold. Research Journal of Pharmacy and Technology. 2022, 15, 419-3. doi:10.52711/0974-360X.2022.00069.
CrossRef - Patel, H.M.; Pawara, R.; Ansari, A.; Noolvi, M.; Surana, S. Design and synthesis of quinazolinones as EGFR inhibitors to overcome EGFR resistance obstacle. Bioorg Med Chem. 2017, 25, 2713-2723. doi: 10.1016/j.bmc.2017.03.039.
CrossRef - Constantinescu, T.; Lungu, C.N. Anticancer Activity of Natural and Synthetic Chalcones. Int J Mol Sci. 2021,22 ,11306. doi: 10.3390/ijms222111306
CrossRef - Jung, S.K.; Lee, M.H.; Lim, D.Y.; Kim, J.E.; Singh, P.; Lee, S.Y.; Jeong, C.H.; Lim, T.G.; Chen, H.; Chi, Y.I.; Kundu, J.K.; Lee, N.H.; Lee, C.C.; Cho, Y.Y.; Bode, A.M.; Lee, K.W.; Dong, Z. Isoliquiritigenin induces apoptosis and inhibits xenograft tumor growth of human lung cancer cells by targeting both wild type and L858R/T790MM mutant EGFR. J Biol Chem. 2014, 289, 35839-48. doi: 10.1074/jbc.M114.585513.
CrossRef - Zhuang, C.; Zhang, W.; Sheng, C.; Zhang, W.; Xing, C.; Miao, Z. Chalcone: A privileged structure in medicinal chemistry. Chem Rev. 2017, 117, 7762-10. doi: 10.1021/acs.chemrev.7b00020.
CrossRef - Shalini, K.V. Have molecular hybrids delivered effective anti-cancer treatments and what should future drug discovery focus on? Expert Opin Drug Discov. 2021,16,335-363. doi: 10.1080/17460441.2021.1850686
CrossRef - Singh, A.K.; Kumar, A.; Singh, H.; Sonawane, P.; Paliwal, H.; Thareja, S.; Pathak, P.; Grishina, M.; Jaremko, M.; Emwas, A.H.; Yadav, J.P.; Verma, A.; Khalilullah, H.; Kumar, P. Concept of Hybrid Drugs and Recent Advancements in Anticancer Hybrids. Pharmaceuticals (Basel). 2022,15, 1071. doi: 10.3390/ph15091071.
CrossRef - Wani, Z.A.; Guru, S.K.; Rao, A.V.; Sharma, S.; Mahajan, G.; Behl, A.; Kumar, A.; Sharma, P.R.; Kamal, A.; Bhushan, S.; Mondhe, D.M. A novel quinazolinone chalcone derivative induces mitochondrial dependent apoptosis and inhibits PI3K/Akt/mTOR signaling pathway in human colon cancer HCT-116 cells. Food Chem Toxicol. 2016, 87,1-11. doi: 10.1016/j.fct.2015.11.016.
CrossRef - Madhavi, S.; Sreenivasulu, R.; Yazala, J.P.; Raju, R.R. Synthesis of chalcone incorporated quinazoline derivatives as anticancer agents. Saudi Pharm J. 2017, 25,275-279. doi: 10.1016/j.jsps.2016.06.005.
CrossRef - Abbas, S.H.; Abd El-Hafeez, A.A.; Shoman, M.E.; Montano, M.M.; Hassan, H.A. New quinoline/chalcone hybrids as anti-cancer agents: Design, synthesis, and evaluations of cytotoxicity and PI3K inhibitory activity. Bioorg Chem. 2019, 82,360-377. doi: 10.1016/j.bioorg.2018.10.064.
CrossRef - Zhou, W.; Zhang, W.; Peng, Y.; Jiang, Z.H.; Zhang, L.; Du, Z. Design, Synthesis and Anti-Tumor Activity of Novel Benzimidazole-Chalcone Hybrids as Non-Intercalative Topoisomerase II Catalytic Inhibitors. Molecules. 2020;25, 3180. doi: 10.3390/molecules25143180.
CrossRef - Kasetti, A.B.; Singhvi, I.; Nagasuri, R.; Bhandare, R.R.; Shaik, A.B. Thiazole-Chalcone Hybrids as Prospective Antitubercular and Antiproliferative Agents: Design, Synthesis, Biological, Molecular Docking Studies and In Silico ADME Evaluation. Molecules. 2021; 26,2847. doi: 10.3390/molecules26102847.
CrossRef - Alam, M.J.; Alam, O.; Perwez, A.; Rizvi, M.A.; Naim, M.J.; Naidu, V.G.M.; Imran, M.; Ghoneim, M.M.; Alshehri, S.; Shakeel, F. Design, Synthesis, Molecular Docking, and Biological Evaluation of Pyrazole Hybrid Chalcone Conjugates as Potential Anticancer Agents and Tubulin Polymerization Inhibitors. Pharmaceuticals (Basel).2022,15,280. doi: 10.3390/ph15030280.
CrossRef - Bao, S.M.; Hu, Q.H.; Yang, W.T.; Wang, Y.; Tong, Y.P.; Bao, W.D. Targeting Epidermal Growth Factor Receptor in Non-Small-Cell-Lung Cancer: Current State and Future Perspective. Anticancer Agents Med Chem. 2019,19, 984-991. doi: 10.2174/1871520619666190313161009.
CrossRef - Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020,5,22. doi: 10.1038/s41392-020-0116-z.
CrossRef - Zhang, H. Three generations of epidermal growth factor receptor tyrosine kinase inhibitors developed to revolutionize the therapy of lung cancer. Drug Des Devel Ther.2016 ,10:3867-3872. doi: 10.2147/DDDT.S119162.
CrossRef - Troiani, T.; Napolitano, S.; Della, C.M.; Martini, G.; Martinelli, E.; Morgillo, F.; Ciardiello, F. Therapeutic value of EGFR inhibition in CRC and NSCLC: 15 years of clinical evidence. ESMO Open. 2016,1,e000088. doi: 10.1136/esmoopen-2016-000088.
CrossRef - Sequist, L.V.; Yang, J.C.; Yamamoto, N.; O’Byrne, K.; Hirsh,V; Mok, T; Geater, S.L.; Orlov, S.; Tsai, C.M.; Boyer, M.; Su, W.C.; Bennouna, J.; Kato, T.; Gorbunova, V.; Lee, K.H.; Shah, R.; Massey, D.; Zazulina, V.; Shahidi, M.; Schuler, M. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013, 31,3327–3334. doi: 10.1200/JCO.2012.44.2806.
CrossRef - Cross, D.A.; Ashton, S.E.; Ghiorghiu, S.; Eberlein, C.; Nebhan, C.A.; Spitzler, P.J.; Orme, J.P.; Finlay, M.R.; Ward, R.A.; Mellor, M.J.; Hughes, G.; Rahi, A.; Jacobs, V.N.; Red Brewer, M.; Ichihara, E.; Sun, J.; Jin, H.; Ballard, P.; Al-Kadhimi, K.; Rowlinson, R.; Klinowska, T.; Richmond, G.H.; Cantarini, M.; Kim, D.W.; Ranson, M.R.; Pao, W. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014, 4,1046–1061. doi: 10.1158/2159-8290.CD-14-0337.
CrossRef - Zhou, J.; Ji, Q.; Li, Q. Resistance to anti-EGFR therapies in metastatic colorectal cancer: underlying mechanisms and reversal strategies. J Exp Clin Cancer Res. 2021,40 ,328. doi: 10.1186/s13046-021-02130-2.
CrossRef - Kargbo, R.B. Targeting the KRAS G12D Mutant as Potential Therapy in Cancer. ACS Med Chem Lett. 2021,12, 1212-1213. doi: 10.1021/acsmedchemlett.1c00390.
CrossRef - Sun, Q.; Burke, J.P.; Phan, J.; Burns, M.C.; Olejniczak, E.T.; Waterson, A.G.; Lee, T.; Rossanese, O.W.; Fesik, S.W. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012, 51, 6140-3. doi: 10.1002/anie.201201358.
CrossRef - Bain, D.I.; Smalley, R.K.; Synthesis of 2-substituted-4H-3,1-benzoxazin-4-ones. J. Chem. Soc. C. 1968, 1593-1597.
CrossRef - Gupta,V.D.; Singh, J.; Kinger, M.; Arora A.K.; Jaswal,V.S. Synthesis and Antiviral Activities of Some 2,3-Disubstituted Quinazoline Derivatives. Asian J. Chem. 2015, 27, 4379-4382. doi:10.14233/ajchem.2015.19132
CrossRef - Habib, O.M.O.; Hassan, H.M.; Mekabaty, A.E. Novel quinazolinone derivatives: synthesis and antimicrobial activity. Med Chem Res. 2013, 22,507–519. doi: 10.1007/s00044-012-0079-x.
CrossRef - Wang, Y.T.; Qin, Y.J.; Zhang, Q.Y.L.; Li, U.Y.; Rao, B.; Zhang, Y.Q.;Yang, M.R.; Jiang, A.Q.; Qi, J.L.; Zhu, H.L. Synthesis, biological evaluation, and molecular docking studies of novel chalcone oxime derivatives as potential tubulin polymerization inhibitors.RSC Adv. 2014, 4,32263-32275.
CrossRef - Desai,V.; Desai,S.; Gaonkar,S.N.; Palyekar,U.; Joshi, S.D.; Dixit, S.K. Novel quinoxalinyl chalcone hybrid scaffolds as enoyl ACP reductase inhibitors: Synthesis, molecular docking and biological evaluation. Bioorg Med Chem Lett. 2017, 27,2174-2180. doi: 10.1016/j.bmcl.2017.03.059.
CrossRef - Kurokawa, M.; Wadhwani, A.; Kai, H.; Hidaka, M.; Yoshida, H.; Sugita, C.; Watanabe, W.; Matsuno, K.; Hagiwara, A. Activation of Cellular Immunity in Herpes Simplex Virus Type 1-Infected Mice by the Oral Administration of Aqueous Extract of Moringa oleifera Lam. Leaves. Phytother Res. 2016, 30,797-804. doi: 10.1002/ptr.5580.
CrossRef - El-Hachem, N.; Haibe-Kains, B.; Khalil, A.; Kobeissy, F.H.; Nemer, G. AutoDock and AutoDockTools for Protein-Ligand Docking: Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1(BACE1) as a Case Study. Methods Mol Biol. 2017, 1598:391-403. doi: 10.1007/978-1-4939-6952-4_20.
CrossRef - Bitencourt-Ferreira, G.; Pintro, V.O.; de Azevedo, W.F. Jr. Docking with AutoDock4. Methods Mol Biol. 2019, 2053,125-148. doi: 10.1007/978-1-4939-9752-7_9.
CrossRef - Hirano, T.; Yasuda, H.; Hamamoto, J.; Nukaga, S.; Masuzawa, K.; Kawada, I.; Naoki, K.; Niimi, T.; Mimasu, S.; Sakagami, H.; Soejima, K.; Betsuyaku, T. Pharmacological and Structural Characterizations of Naquotinib, a Novel Third-Generation EGFR Tyrosine Kinase Inhibitor, in EGFR-Mutated Non-Small Cell Lung Cancer. Mol Cancer Ther. 2018, 17,740-750. doi: 10.1158/1535-7163.MCT-17-1033.
CrossRef - Le, Y.; Gan, Y.; Fu, Y.; Liu, J.; Li,W.; Zou, X.; Zhou, Z.; Wang, Z.; Ouyang, G.; Yan, L. Design, synthesis and in vitro biological evaluation of quinazolinone derivatives as EGFR inhibitors for antitumor treatment. J Enzyme Inhib Med Chem. 2020, 35,555-564. doi: 10.1080/14756366.2020.1715389.
CrossRef - Zhang,Y.; Wang, Q.; Li, L.; Le, Y.; Liu, L.; Yang, J.; Li, Y.; Bao, G.; Yan, L. Synthesis and preliminary structure-activity relationship study of 3-methylquinazolinone derivatives as EGFR inhibitors with enhanced antiproliferative activities against tumour cells. J Enzyme Inhib Med Chem. 2021, 36,1205-1216. doi: 10.1080/14756366.2021.1933466.
CrossRef - Agema, B.C.; Veerman; G.D.M.; Steendam, C.M.J.; Lanser, D.A.C.; Preijers, T.; Van der Leest, C.; Koch, B.C.P.; Dingemans, A.C.; Mathijssen, R.H.J.; Koolen, S.L.W. Improving the tolerability of osimertinib by identifying its toxic limit. Ther Adv Med Oncol. 2022,14,1-10. doi: 10.1177/17588359221103212.
CrossRef - Li, L.; Zhao, H.; Peng, X.; Liu, J.; Mai, R.; Chen, J.; Lin, L.; Chen, T.; Yan. J.; Shi, J.; Chen, J. Discovery of novel Quinazoline-based KRAS G12C inhibitors as potential anticancer agents. Bioorg Med Chem. 2022,71,116962. doi: 10.1016/j.bmc.2022.116962.
CrossRef - Zuberi, M.; Khan. I.; O’Bryan, J.P. Inhibition of RAS: proven and potential vulnerabilities. Biochem Soc Trans. 2020, 48, 1831-1841. doi: 10.1042/BST20190023.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.









